Gut makeup could make diarrhea less likely
Molecular signature of a diarrhea-resistant gut found
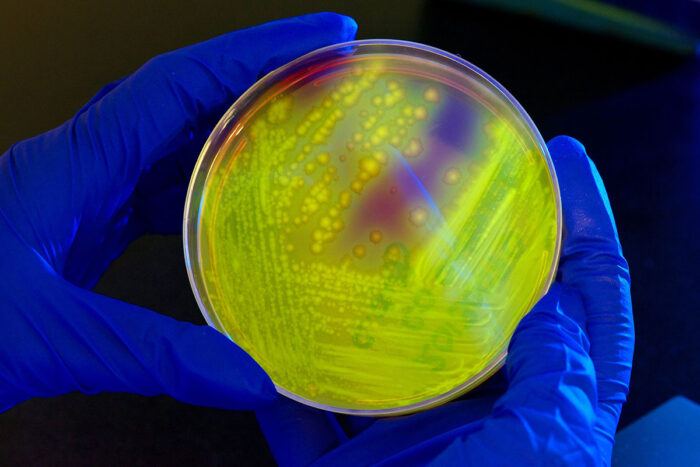 CDC/Melissa Dankel
CDC/Melissa DankelThe intestinal bacteria Clostridium difficile (C. difficile) glows yellow-green on a petri dish when illuminated by UV light. Researchers at Washington University School of Medicine in St. Louis have found the molecular signature of a healthy gut microbial community – the kind that can keep C. difficile from overgrowing and causing diarrhea.
Antibiotics are known to upset the balance of bacteria in the intestinal tract. In some cases, antibiotics can cause the bacterium Clostridium difficile (C. difficile) to overgrow wildly, causing diarrhea and, in severe cases, life-threatening intestinal inflammation.
Researchers at Washington University School of Medicine in St. Louis have found the molecular signature of a healthy gut microbial community, or microbiome – the kind of community that keeps C. difficile in check even in the aftermath of antibiotic treatment. They also have identified a specific molecule produced when C. difficile is not lying dormant but is active and making toxins. Together, the findings outline a set of molecular signs that indicate a person has – or is at risk for – diarrhea caused by C. difficile.
“Right now, we just accept that taking antibiotics raises the risk of C. diff infection,” said senior author Jeffrey P. Henderson, MD, PhD, an associate professor of medicine and of molecular microbiology. “By analyzing the small molecules produced by the microbiome, we may be able to identify people at high risk for developing C. diff diarrhea. We also may be able to use this type of analysis to screen potential donors for fecal transplants and make sure they are donating the kind of microbiome that can help keep C. diff under control.”
The findings are published Aug. 12 in The Journal of Clinical Investigation.
Every year, there are about 450,000 cases of C. difficile diarrhea in the U.S., and 29,000 deaths. Henderson and colleagues – including first author and postdoctoral researcher John Robinson, PhD, and co-author Erik Dubberke, MD, a professor of medicine, both at Washington University; as well as co-author Peter Mucha, PhD, a professor of mathematics at the University of North Carolina at Chapel Hill – studied the microbiomes of people with and without C. difficile disease by analyzing their metabolomes, or the chemical compounds produced in the gut as human intestinal cells and microbes eat, breathe and interact with each other. They were searching for a molecular signature that distinguishes a healthy gut from one prone to C. difficile disease.
The search was complicated by the fact that some people carry C. difficile in their gastrointestinal tracts with no ill effects. But the bacteria also can produce two toxins that cause diarrhea and inflammation. In the absence of these toxins, the bacteria are mostly harmless.
The researchers studied 186 hospitalized people with diarrhea, divided into three groups: a group with no C. difficile; a group with C. difficile but without toxins, which means they carried the bacteria but their symptoms were caused by something else; and a group with C. difficile and toxins, all of whom had C. difficile infections.
Using mass spectrometry, the researchers analyzed thousands of metabolic byproducts in stool samples from patients in all three groups. In particular, they found high levels of a fatty acid called 4-methylpentanoic acid in the stool of people with C. difficile disease. The fatty acid is produced when proteins are broken down for fuel using an unusual metabolic process. Human cells don’t produce the fatty acid, and neither do most of the bacteria in the normal microbiome – except for C. difficile. The presence or absence of the fatty acid in stool identified people with C. difficile disease with 92.8 percent accuracy.
Further, the researchers uncovered a pattern of molecules related to bile acid metabolism that was linked to protection against C. difficile disease. Bile acids are produced by the liver to help with fat digestion, then absorbed into the intestine, where members of the microbiome chemically modify them. The researchers identified a set of modified bile acids in people who did not carry the bacterium, or who carried it harmlessly, that was absent from people with C. difficile infections.
“These unusual bile acids may be fingerprints of people who are more resistant to C. difficile infection,” Henderson said. “There seems to be a difference in the metabolism of bile acids by the microbiome that affects how likely people are to develop disease.”
The researchers now are working on identifying which bacteria in the microbiome are involved in producing the protective bile acids, and how a healthy microbiome keeps C. difficile restrained.
“We know that disruptions to the microbiome predispose some people to getting C. diff disease, but until now we haven’t known much about what these changes are and why they’re harmful,” Henderson said. “Small molecules give us a direct readout of what the microbiome is doing. This study provides some big clues as to how these antibiotic-resistant bacteria make people sick, and that could lead to better ways to identify, prevent or treat C. diff infections.”