Online database aims to collect, organize research on cancer mutations
Goal is to help clinicians treat patients based on tumor genetics
 Joshua McMichael
Joshua McMichaelAn online "knowledgebase" called CIViC is an open access resource for collecting and interpreting information from the vast body of scientific publications on cancer genetics. The hope is this "Wikipedia" of cancer genomics will help doctors develop precision medical therapies for patients with cancer, according to researchers at Washington University School of Medicine in St. Louis.
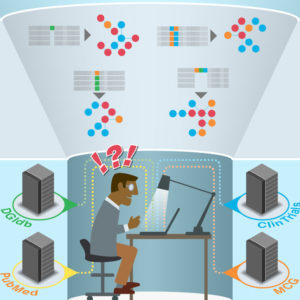
The body of knowledge on cancer genomics is massive and ever-expanding. But this wealth of potentially critical information is far less likely to be of help to patients if it is inaccessible to the doctors who treat them.
Researchers at Washington University School of Medicine in St. Louis have developed an online “knowledgebase” intended for the gathering and organization of this information so that clinicians have improved chances of identifying important mutations in a patient’s tumor and potentially connecting genetic errors with drugs known to target them.
The online resource, called CIViC, is described Jan. 30 in Nature Genetics.
CIViC stands for Clinical Interpretations of Variants in Cancer, and the researchers liken it to a Wikipedia of cancer genetics. Anyone can create an account and contribute information. That information is then curated by editors and moderators who are experts in the field.
“It’s relatively easy now to sequence the DNA of tumors — to gather the raw information — but there’s a big interpretation problem,” said senior author Obi L. Griffith, PhD, an assistant professor of medicine. “What do these hundreds or thousands of mutations mean for this patient? There are a lot of studies being done to answer these questions. But oncologists trying to interpret the raw data are faced with an overwhelming task of plumbing the literature, reading papers, trying to understand what the latest studies tell them about these mutations and how they may or may not be important.”
The CIViC knowledgebase is an attempt to solve this problem. While the investigators said this is one of many efforts to collect and interpret such information, CIViC is the only one that is entirely open access, to their knowledge. Anyone is free to contribute and use the content as well as the source code.
“We are committed to keeping this resource open and available to anyone who wants to contribute or make use of the information,” said Malachi Griffith, PhD, an assistant professor of medicine. “We would like it to be a community exercise and public resource. The information is in the public domain. There are no restrictions on its use, academic or commercial.”
Obi and Malachi Griffith, who are twin brothers, are assistant directors of the university’s McDonnell Genome Institute.
Though anyone can submit a new piece of information or suggest edits to existing data, at least two independent contributors must agree that the new information should be incorporated, and one of those users must be an expert editor. Expert editors are not permitted to approve their own submissions. Information on the CIViC website provides details about how new users may be promoted to expert editors and administrators.
To date, the site has seen over 17,500 users from academic institutions, governmental organizations and commercial entities around the world. Since CIViC’s launch, 59 users have volunteered their time to contribute their knowledge to CIViC, including descriptions of the clinical relevance of 732 mutations from 285 genes for 203 types of cancer, all gleaned from reviewing 1,090 scientific and medical publications.
Despite the fact there are many groups attempting to collect and interpret genomic variants in cancer, the investigators said the sheer volume of information has resulted in relatively little overlap in data gathered so far.
“While we believe this is the only such open-access knowledgebase, there are other large research centers with similar resources,” Malachi Griffith said. “We did an analysis to compare the big ones. Even though we all have access to the same published literature, if you look at the overlap of the information mined by each of these resources, it’s remarkably small. We’re all approaching the same problem and just by chance — and probably because of the amount of information out there — we haven’t duplicated our efforts very much yet.”
Obi and Malachi Griffith said finding a way to combine these resources is the primary goal of an international group they are helping lead called the Variant Interpretation for Cancer Consortium, which is a part of the Global Alliance for Genomics and Health (GA4GH).
“We’re just scratching the surface of the potential this holds for precision medicine,” Obi Griffith said. “There’s a lot of work to do.”